PowerMV: A software environment for statistical analysis, molecular viewing, descriptor generation, and similarity search.
Developed by :-Jack Liu, Jun Feng, Atina Brooks and Stan Young National Institute of Statistical Sciences.
Prerequisites:
1. Microsoft .net 1.1 and above (required)
3. DirectX 9.0c Runtime (optional for 3D viewing)
Basic Functions:
- Supports MDL SDF format
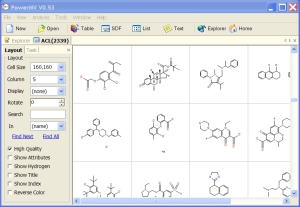
• Displays molecules in multiple columns. - Displays properties contained in SD file in a table.
• Anti-alias technology for best picture quality.
• Table of molecule pictures and properties can be exported to Excel (Office XP and above) to generate personalized reports.
• Calculates three types of binary atom pair descriptors and continuous weighed burden numbers.
• Searches over ACL library to determine possible mechanisms or side effects. The user can create and load their personal databases. - Calculates Drug-like properties like LogP, PSA, MW, HBAs, HBDs, etc.
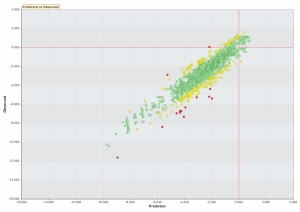
- Builds regression model using Least Angle Regression (LARS) and LASSO-2
- Builds regression and classification model using Random Forest through graphical interface to R.
- Cluster analysis with KMeans through graphical interface to R.
- Outlier detection using tetrads method (Douglas Hawkins, et al). (Code implemented by Andrew Wong).
- Novel robust single value decomposition (RSVD) for large datasets with missing values or outliers.
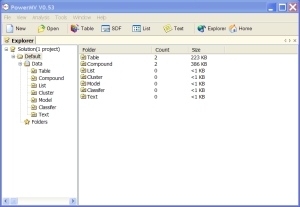
GUI overview: Projects are stored on the left and contents of a folder on the right.
Compound Viewing: Compounds are displayed in a grid. The cell size and number of columns in the display are under user control.